Use Cases
Use Cases¶
- Phenotypic profile matching
- Computational identification of disease models through cross-species phenotype comparison
- Aggregating phenotype data across species
- Predicting phenotype associations using AI
- Cross-species inference in Variant and Gene Prioritisation algorithms (Exomiser).
- Use cases for uPheno in the Alliance of Genome Resources and MGI
- Cross-species data in biomedical knowledge graphs (Kids First)
Phenotypic profile matching¶
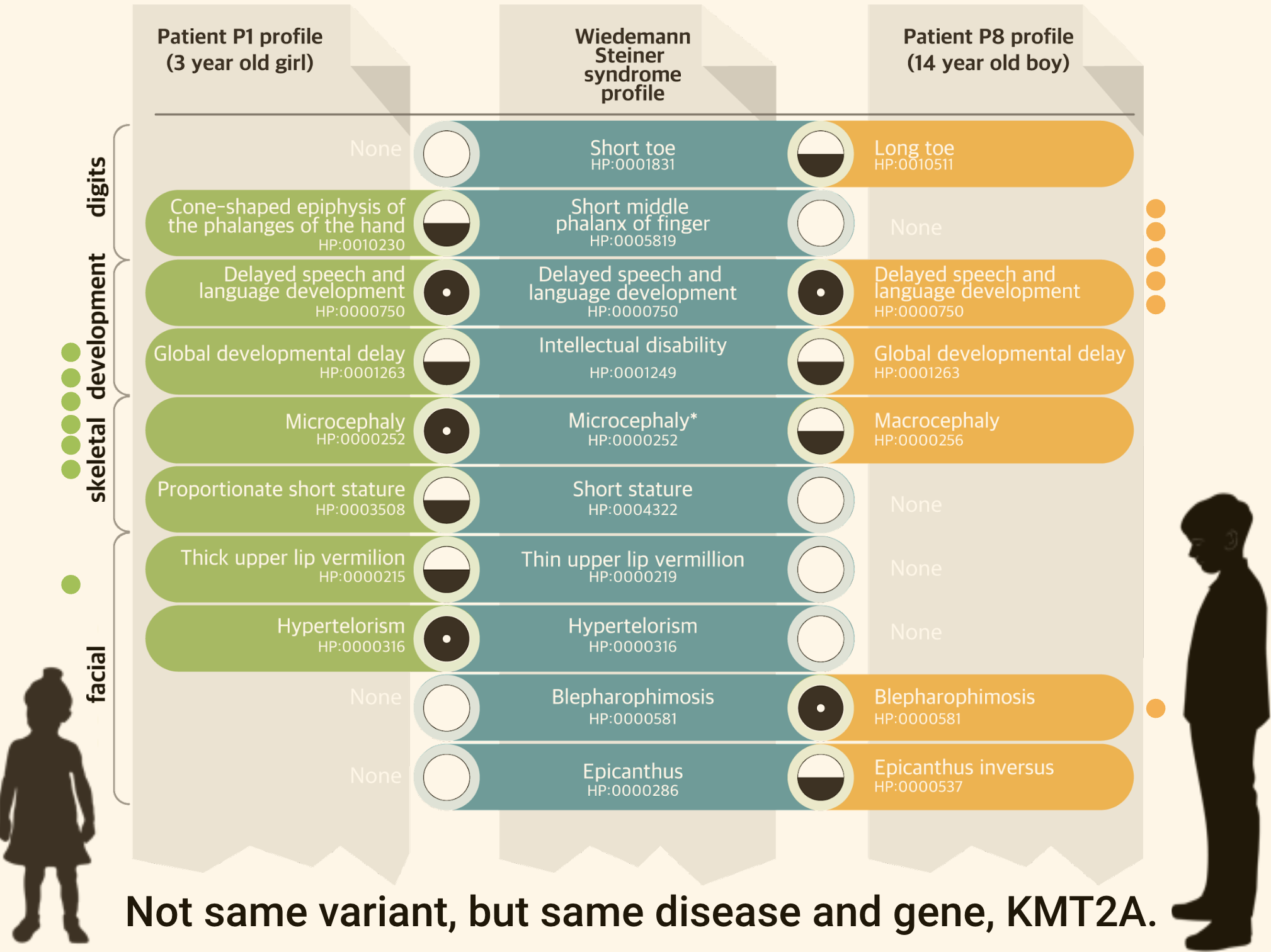
Figure 1: Phenotypic profile matching
Phenotype profiles can be compared to known disease profiles (i.e. the phenotypic features commonly associated with a disease). They can also compared directly to find similar cases, e.g. Matchmake Exchange.
Computational identification of disease models through cross-species phenotype comparison¶
See presentation by Diego Pava during a uPheno stakeholder meeting for details on how IMPC leverages cross-species phenotype inference for disease model identification.
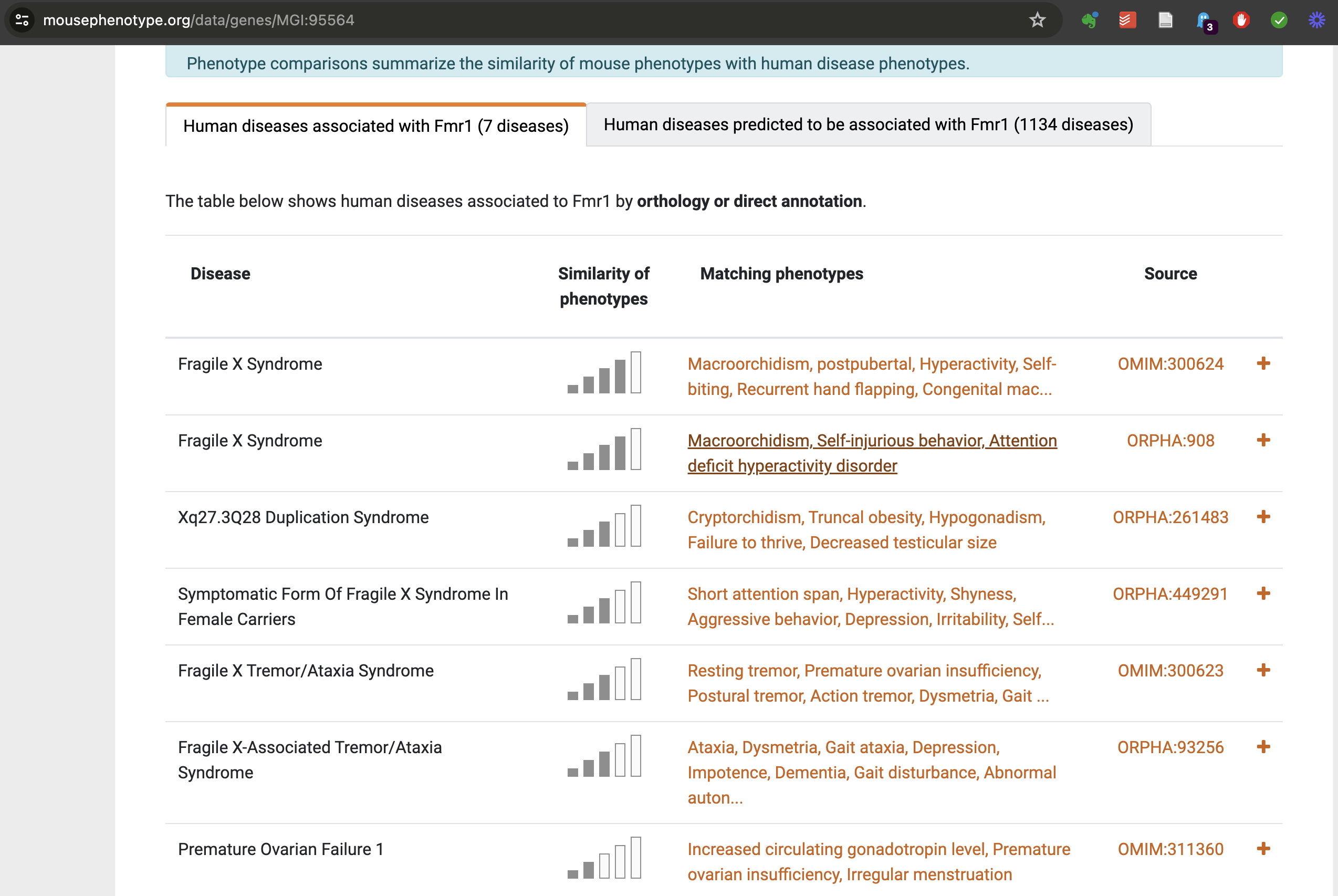
Figure 2: Phenotypic profile matching
Diseases associated with a gene through cross-species inference. Source: https://www.mousephenotype.org/data/genes/MGI:95564.
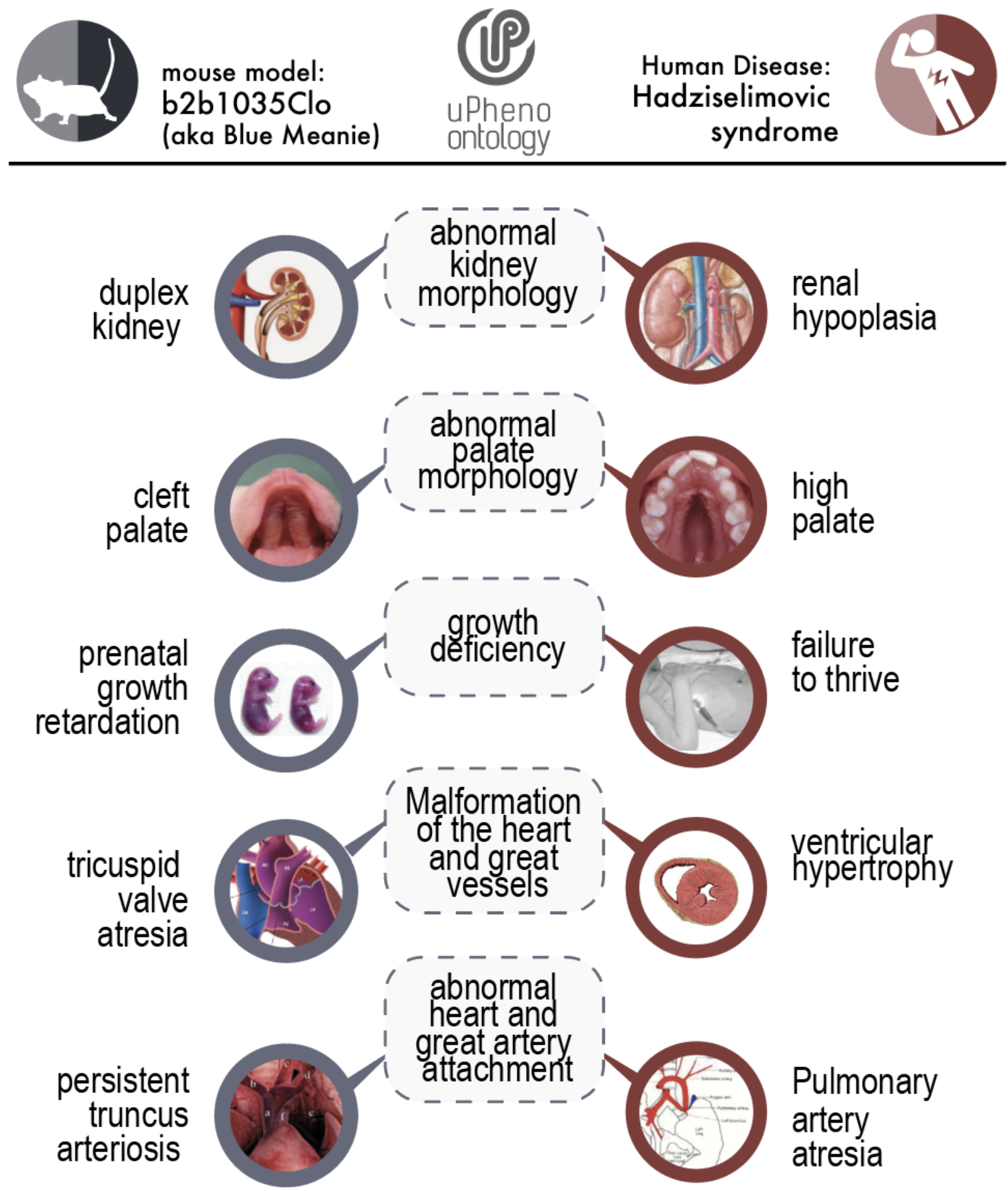
Figure 3: Mouse model comparison with uPheno
A mouse profile on the left is associated with a disease profile on the right through common, species independent subsumers.
Aggregating phenotype data across species¶
See also section on how to integrate phenotype data.
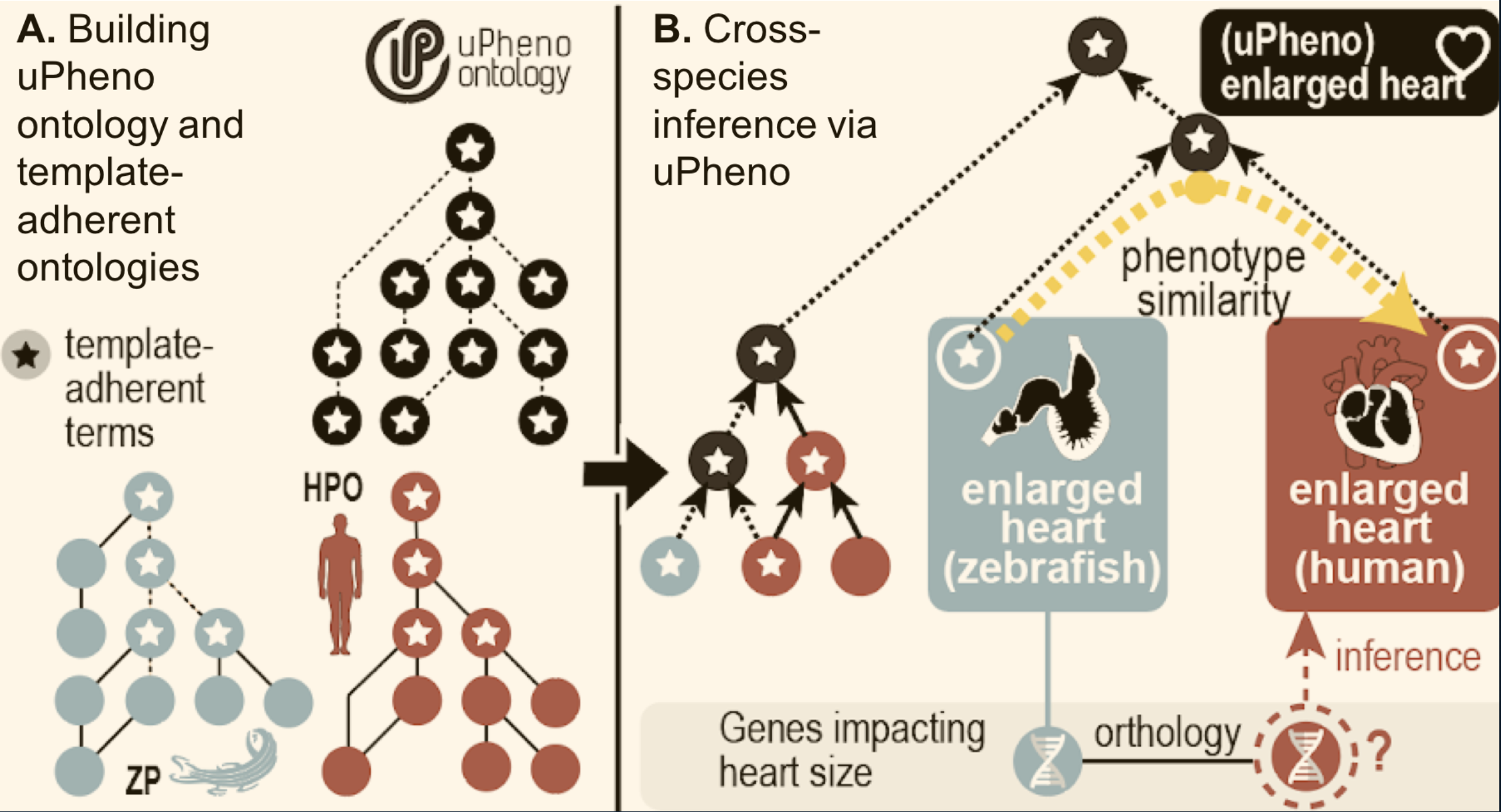
Figure 4: uPheno cross-species integration
uPheno integrates species-specific pre-coordinated phenotype ontologies such as HPO and ZP. Species specific phenotype terms like "enlarged heart (ZP)" or "Enlarged heart (HPO)" are integrated under a common uPheno class which is species-independent.
Predicting phenotype associations using AI¶
TBD.
Cross-species inference in Variant and Gene Prioritisation algorithms (Exomiser)¶
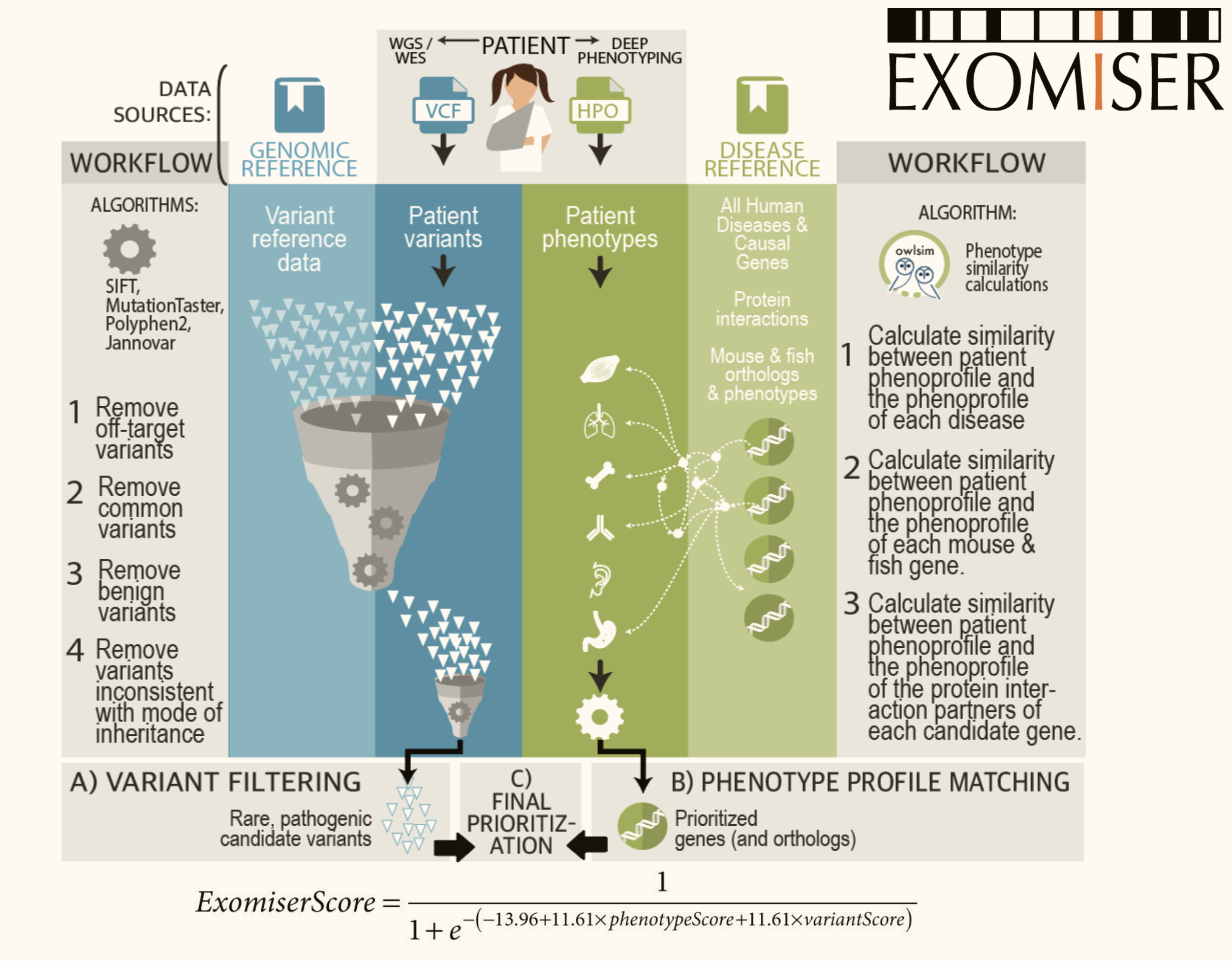
Figure 5: The Exomiser pipeline
Exomiser is a phenotype-powered Variant and Gene Prioritisation System.